The content in this section seeks to provide a clinical summary of information and resources for medical representatives treating 22q patients.
Clinical Summary
Differential Diagnosis
For current information on availability of genetic testing for disorders included in this section, see GeneTests Laboratory Directory. —ED.
Each of the anomalies seen in the 22q11.2 deletion syndrome can be found as an isolated anomaly in an otherwise normal individual.
Up to 8% of individuals with an isolated palatal cleft, including submucosal cleft, may have deletion 22q11.2, making this the most common genetic syndrome associated with palatal clefts. Conversely, the 22q11.2 deletion syndrome is the most common genetic basis of congenital velopharyngeal incompetence.
Disorders with overlapping features:
- Smith-Lemli-Opitz syndrome (when polydactyly and cleft palate are present). Smith-Lemli-Opitz syndrome is associated with elevated serum concentration of 7-dehydrocholesterol (7-DHC) or an elevated 7-dehydrocholesterol:cholesterol ratio. Molecular genetic testing for mutations of the DHCR7 gene is available.
- Alagille syndrome (when butterfly vertebrae, congenital heart disease, and posterior embryotoxon are present). Sequence analysis of the JAG1 gene detects mutations in more than 70% of individuals who meet clinical diagnostic criteria. FISH detects a microdeletion of 20p12, including the entire JAG1 gene, in approximately 5-7% of affected individuals.
- VATER syndrome (when heart disease, vertebral, renal, and limb anomalies are present)
- Oculo-auriculo vertebral (Goldenhar) syndrome (when ear anomalies, vertebral defects, heart disease, renal anomalies are present)
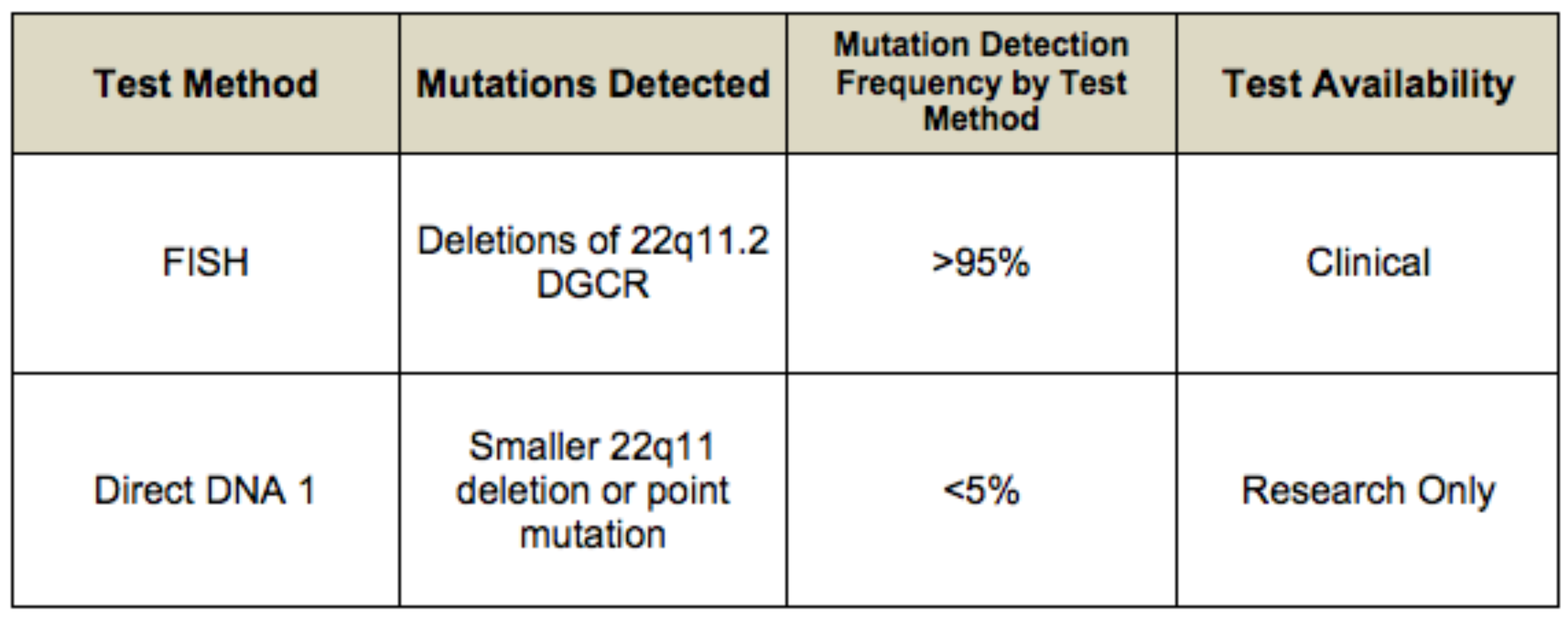
Individuals suspected of having the 22q11.2 deletion syndrome but having normal FISH studies may have a chromosome abnormality involving some other chromosomal region, including deletion 10p13-p14.
Natural History
Findings in 250 individuals (48% male; 52% female) with 22q11.2 deletion syndrome are summarized below [McDonald-McGinn et al 1999b]. In unpublished data on 600 individuals, the percentages for the following findings remain the same [Author, unpublished data 2005].
Thirty-three percent of individuals were five years of age or younger. Marked inter- and intrafamilial variability is observed.
Genotype-Phenotype Correlations
The majority of individuals have the same large deletion of the DGCR. Of note, the size of the deletion remains unchanged with parent-to-child transmission. The great inter- and intrafamilial clinical variability makes genotype-phenotype correlations difficult [Driscoll et al 1995, McDonald-McGinn et al 2001]. Anecdotally, developmental delays appear to be more significant in familial cases; however, this may reflect socioeconomic rather than genetic factors.
Penetrance
Penetrance is complete in individuals with deletion 22q112 detected by FISH; variability is marked.
Anticipation
To date, anticipation has not been observed.
Nomenclature
It is now recognized that the 22q11.2 deletion syndrome encompasses the phenotypes previously called DiGeorge syndrome (DGS), velocardiofacial syndrome (VCFS) (Shprintzen syndrome), conotruncal anomaly face syndrome (CTAF) [Matsuoka et al 1994], many cases of the autosomal dominant Opitz G/BBB syndrome [McDonald-McGinn et al 1995, Fryburg et al 1996, LaCassie & Arriaza 1996], and Cayler cardiofacial syndrome (asymmetric crying facies) [Giannotti et al 1994]. The clinical descriptions of DGS, VCFS, and CTAF resulted from an ascertainment bias.
DGS was originally described as a developmental field defect of the third and fourth pharyngeal pouches with a conotruncal cardiac anomaly and aplasia or hypoplasia of the thymus gland and parathyroid glands. Later, congenital heart disease was added. The majority of individuals with DGS were identified in the neonatal period with a major congenital heart defect, hypocalcemia, and immunodeficiency.
VCFS, also called Shprintzen syndrome, was originally described as the combination of velopharyngeal incompetence (VPI), congenital heart disease (usually a ventricular septal defect or tetralogy of Fallot), characteristic facial features, and developmental delay or learning difficulties. Children with VCFS tended to be diagnosed in cleft palate clinics or craniofacial centers when they reached school age and speech and learning difficulties became evident [Wilson et al 1993, Wulfsberg et al 1996, McDonald-McGinn et al 1997b, Thomas & Graham 1997].
Prevalence
Estimates of prevalence vary from one in 4000 [Wilson et al 1994] to one in 6395 [Devriendt et al 1998]. Given the variable expression of the deletion 22q11.2, the incidence is probably much higher than previously estimated. In a population-based study in Sweden, the mean annual incidence was 14.1 per 100,000 live births [Oskarsdottir et al 2004, Oskarsdottir et al 2005a, Oskarsdottir et al 2005b]. A U.S. population-based study conducted by the Centers for Disease Control (CDC) found an overall prevalence of about one in 6000 in whites, blacks, and Asians, and one in 3800 in Hispanics [Botto et al 2003].